WHAT IS IT?
------------
This model explains kinetics of radical polymerization. Radical polymerization
can be thought of as the process of agglomeration of small molecules (monomers)
into chains (polymer chains) initiated by active particles (radicals).
An example of radical polymerization would be industrial polymerization of
ethylene into polyethylene (PET):
n C H --------> ~C H~
2 4 2n 4n
A gaseous mixture of ethylene and oxygen (used as a catalyst), is pumped through
a mile-long heated pipe. As the mixture moves along the pipe, the molecules of
ethylene polymerize into chains. At the end of the pipe, the polymer is
collected and co-products with low molecular mass are recycled.
Molecules of oxygen turn into bi-radicals under the influence of high
temperature:
O --------> *O-O*
2
Here the asterisk denotes a free electron. These bi-radicals react with pi-bonds
in ethylene and start the process of chain growth according to the scheme:
*O-O* + C H --------> *O-O-CH-CH*
2 4 2 2
*O-O-CH-CH* + CH-CH --------> *O-O-CH-CH-CH-CH*
2 2 2 2 2 2 2 2
We can thus think of radical polymerization as interaction between (bi-) radicals
and monomers.
Notice that if two radicals react with each other, they will form an inert
compound and end the process of forming loner chains.
Let us assume that we run polymerization using AIBN as an initiator which we
denote by I. We thus have the following kinetically distinct particles in the
system:
(i) Initiator molecules; (ii) Radicals; (iiI) Monomers; (Iv) "Dead"
chains;
A radical can react with a monomer, in which case it absorbs the monomer and
increases its chain-length by 1. Alternatively, a radical can react with another
radical forming a "dead" molecule which does not react with other particles. Let
us denote by R(n) radicals which have reacted n times with monomers. Let M stand
for monomers and C(k) for "dead" chains of length k. Then schematically we can
describe the reactions by the following system of equations:
I --------> 2 R(1) (initiation)
R(n) + M --------> R(n+1) (growth)
R(n) + R(m) --------> C(n+m) (termination)
In this model, there are four breeds of turtles:
-initiator-breed (yellow turtles);
-monomer-breed (green turtles);
-radical-breed (red turtles);
-polymer-breed (blue turtles).
Each turtle carries two variables: chain-length and id. Initially there are 2000
turtles of monomer breed with id set to "nobody" and chain-length set to 1.
There is also a user-defined number (Io) of initiator breed with id set to
"nobody" and chain-length 0.
The rules are quite simple:
(i) All turtles move around randomly. However no turtle can go to a blue
patch. (Think of blue patches as of glass walls of a test tube.) (ii) Yellow
turtles (initiators) spontaneously break up into two red turtles (radicals) with
probability Ki; (iii) Green turtles (monomers) react with red turtles (radicals)
with probability Km. If there is at least one radical on the same patch as the
given monomer, the monomer dies and the chain-length of the radical which
"killed" it is incremented by 1; (iv) Red turtles (radicals) react with each
other pairwise. If there is a pair of radicals on the same patch, they
annihilate each other with probability Kr forming a polymer chain (a blue
turtle);
HOW TO USE IT
-------------
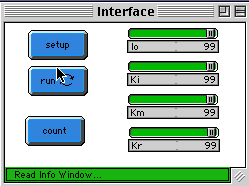
With sliders one can control the initial number of initiator-breed (Io) and
reaction probabilities Ki (initiator): probability of an initiator (yellow)
creating two radicals (red); Km (monomer): probability of reaction between
momomers (green) and radicals (red), forming a radical with a longer chain
length; Kr (radical): probability of two radicals (red) reacting to form a
polymer chain (blue).
Select the initial number of initiator-breed Io and press the "setup" button. To
start the simulation press the "run" button. You can set the probabilities Ki,
Km and Kr before or after you do setup. You can also modify this constants on
the fly during the execution of "run" procedure.
The observer will plot the number of initiators, monomers and radicals. You can
view the distribution of chain-length if you press "statistics" button. This
will periodically stop the "run" button and cause the distribution to appear in
the plot window number 4 titled "Distribution." You can program the
distribution to be re-plotted every 100 or so tics using the "every" primitive if
you want more dynamics.
THINGS TO NOTICE
----------------
Distribution of chain-length drastically depends on the mode of initiation. If
you select low values of Io and Ki, then the life expectancy of radicals will
rise leading to longer chains. However in this case the simulation will have to
run too long in order to produce a sizable amount of long-length chains. If you
start the simulation with a lot of initiator which decomposes rapidly, radicals
will tend to react with themselves and form low molecular oligomers.
THINGS TO TRY
-------------
Try to find the optimum values for which the distribution is tight and the
simulation runs at a reasonable pace.
EXTENDING THE MODEL
-------------
Try to implement the model of industrial production of polyethylene (PET)
described above. Are you going to have "dead" chains in this model?
STARLOGOT FEATURES
------------------
Take note of the implementation of the "to initiate" and "to terminate"
procedures listed below.
to initiate if (random 1000) > (1000 - Ki) [ setbreed radical-breed setcolor red
hatch [hatch [move]] die] end
to terminate if (count-radical-breed-here > 1) and ((random 1000) > 999 - Kr) and
(id = nobody) [ setid one-of-radical-breed-here ifelse (id-of id) != nobody
[setid nobody][setid-of id who]]
if id != nobody [ setbreed polymer-breed setcolor blue if who < id
[setchain-length chain-length + chain-length-of id setchain-length-of id
chain-length]] end
Notice how if (random 1000) > (1000 - K) condition translates an integer K into a
probability that the action is executed. This is all it takes to implement
unimolecular kinetics of initiation!
Also, notice that one has to be rather careful with implementing bimolecular
kinetics. In the listing of the "to terminate" procedure, the first IF statement
guarantees that agents will react pairwise, and the second IF statement is needed
to set up the chain-length of the polymer breed correctly.
REFERENCES AND RELATED MODELS
-----------------------------
Also see the model "Kinetics of A + A <=> B"
Although oxygen is widely used in industry as initiator of radical
polymerization, scientists prefer to use other initiators in the lab, especially
when the reaction has to be performed under conidions of atmospheric pressure and
room temperature. Two of the most frequently used lab initiators are benzoyl
peroxide and 2-2'-azo-bis-isobutyrylnitrile (AIBN):
C H-C(O)O-O(O)C-H C
6 5 5 6
CH CH
| 3 | 3
CH -C--N=N-C- CH
3 | | 3
CN CN
Peroxydes and azo-compounds are extremely unstable and tend to break up into
radicals very easily if exposed to moderate heat or light. Here is the reaction
of decomposition of AIBN:
CH CH CH
| 3 | 3 | 3 ^
CH -C--N=N-C- CH --------> 2 CH -C* + N |
3 | | 3 3 | 2
CN CN CN
This reaction is irreversible because of the escape of nitrogen.
